PoseView
The tool PoseView [1] automatically generates two-dimensional diagrams of complexes with known three dimensional structures while adhering to conventions of chemical structure diagram generation.
In a complex diagram hydrogen bonds are represented as dashed lines and both the interacting amino acids of the receptor and the ligand are represented as structure diagram. Hydrophobic contacts between the ligand and protein are represented by spline segments which mark hydrophobic regions of the ligand and the corresponding amino acid descriptors of the PDB-database. The coordinates of the structure diagram and their layout modifications are generated by the library SDGlib [2]. The selection of the shown amino acids is based on the interaction calculations which is done by the FlexX-library[3].
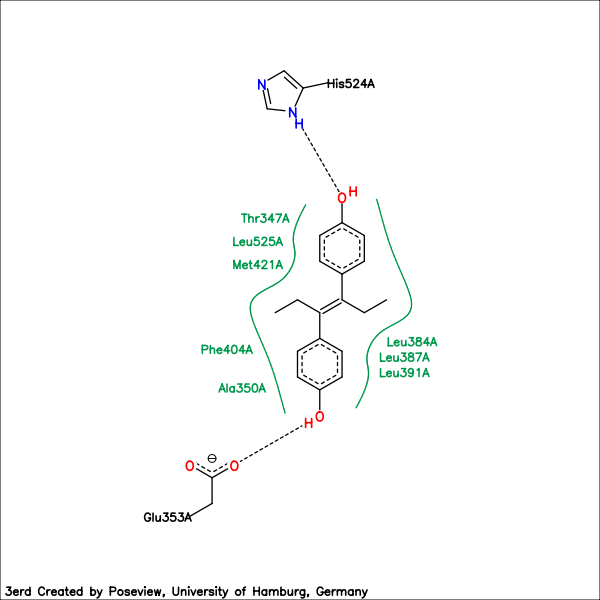 Complex of Estrogen Receptor Alpha and Diethylstilbesterol. PDB code: 3erd.
Complex of Estrogen Receptor Alpha and Diethylstilbesterol. PDB code: 3erd.
PoseView Interface
Features
- Complex diagrams can be shown in a browser or written to a file in one of the following formats: PNG, PDF, SVG or EPS.
- Interacting amino acids can be displayed as structure diagrams of complete molecules or of the interacting molecule part.
- Hydrogen bonds can be labeled by their energy or length.
- Hydrophobic contacts can be hidden.
Referenzen
[1] Stierand, K., Maaß, P., Rarey, M. (2006) Molecular Complexes at a Glance: Automated Generation of two-dimensional Complex Diagrams. Bioinformatics, 22, 1710-1716.
[2] Fricker, P., Gastreich, M., and Rarey, M. (2004) Automated Generation of Structural Molecular Formulas under Constraints. Journal of Chemical Information and Computer Sciences, 44, 1065-1078.
[3] Rarey, M., Kramer, B., Lengauer, T. and Klebe, G. (1996) A fast flexible docking method using an incremental construction algorithm, Journal of Molecular Biology, 261, 470-489.
Stierand, K., Rarey, M. (2007). From Modeling to Medicinal Chemistry: Automatic Generation of Two-Dimensional Complex Diagrams. ChemMedChem 2,6, 853-860
