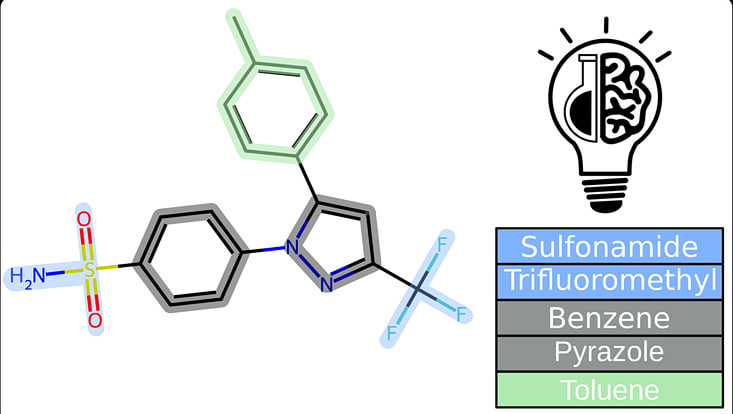
New Paper: LifeSoaks: a tool for analyzing solvent channels in protein crystals and obstacles for soaking experiments
31 August 2023
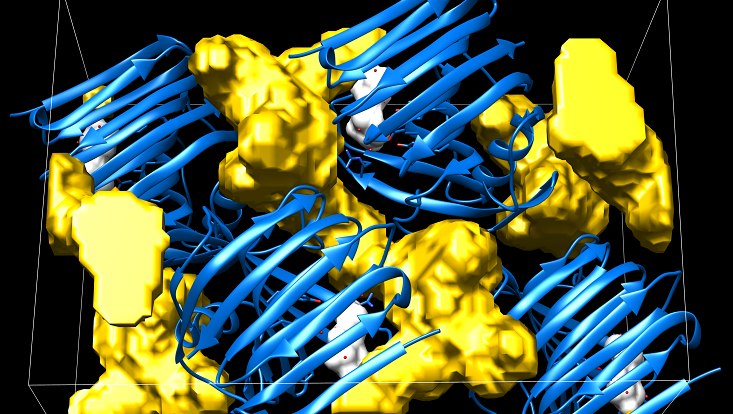
Photo: Jonathan Pletzer-Zelgert
Due to the geometric complexity of protein structures, solvent-filled channels and cavities are omnipresent in their crystal arrangements. These channels are of interest for crystallographers, especially when performing soaking experiments, in which small molecules are expected to diffuse through the preformed crystal and reach a potential binding site.
Our novel Voronoi diagram-based tool, LifeSoaks, enables the computation and visualization of these channels with respect to the size-excluding bottleneck positions on the diffusion paths. We further introduce a hand-curated dataset of successfully soaked structures that has been used to benchmark the performance of LifeSoaks.
Check out the publication for a detailed method description and various application scenarios (https://journals.iucr.org/d/issues/2023/09/00/nz5014/). LifeSoaks can be assessed through our ProteinsPlus webserver (https://proteins.plus) or downloaded from our NAOMI ChemBio software collection at https://software.zbh.uni-hamburg.de