New Paper: Database-Driven Identification of Structurally Similar Protein-Protein Interfaces
14 June 2024
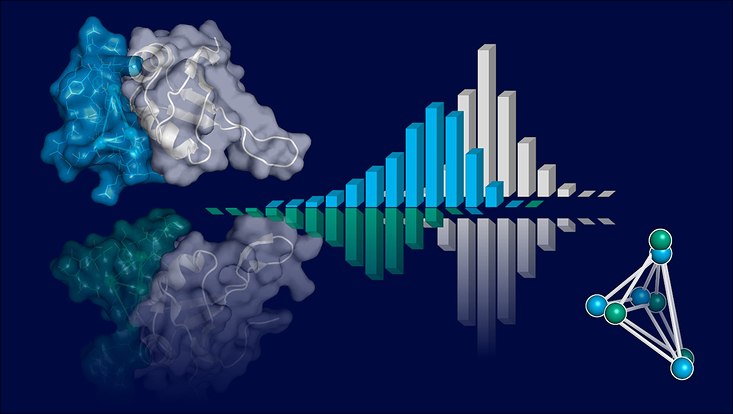
Photo: Christiane Ehrt
Examining the similarity between interfaces of protein-protein complexes can provide valuable insights into protein functions and assist in drug discovery. Typically, tools that measure similarity focus on interactions between two protein interfaces. However, only one predicted interface is available if the task is to discover potential protein binding partners. In this context, we introduce PiMine, a database-driven tool for protein interface similarity search. PiMine compares the interface residues of one or two interacting protein chains by analyzing tetrahedral geometric patterns of α-carbon atoms and calculating physicochemical and shape-based similarities. Using an objective and similarity representation-independent tailor-made dataset, we demonstrate that PiMine surpasses commonly used comparison tools in terms of early enrichment, particularly for interfaces of proteins that are unrelated in sequence and global structure. Additionally, we illustrate its practical application in predicting protein interaction partners by comparing predicted interfaces to known protein-protein interfaces.
Check out the publication for a detailed method description and various application scenarios.
PiMine is available from our NAOMI ChemBio Suite software collection at https://uhh.de/naomi