Mona Documentation
MONA - Common Tasks
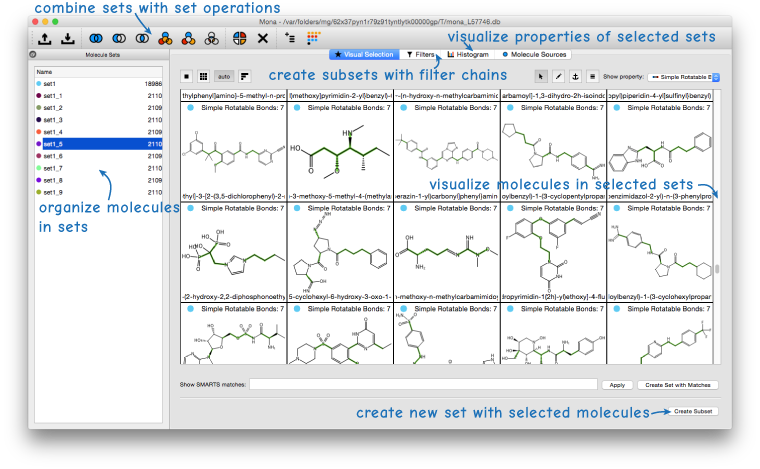
Mona is an interactive tool that can be used to prepare and visualize large collections of small molecules. A set centric workflow allows to intuitively handle hundred thousands of molecules. The following tasks show solutions for typical cheminformatics problems step by step.
Task 1: Which molecules are in my file?
The first task shows how to import molecules into MONA and how to add properties to molecules and visualize them.
1. Click on the Molecule Sets view and select a file to import.
There are multiple ways to import molecules into MONA's internal database (all lead to identical results): Choosing File→Add Molecules.., clicking ![]() , using the Molecule Sources view or clicking in an empty Molecule Sets view.
, using the Molecule Sources view or clicking in an empty Molecule Sets view.
2. Choose which instances are considered identical by MONA.
MONA always handles topologically equal instances as identical. In this dialog you may choose to additionally ignore stereo centers definitions, to treat all tautomers as equal or to neutralize all instances, i.e. ignore any charges on the instances.
3. Double click on the warning  or error
or error  sign in the Molecule Sources view to open the Log view.
sign in the Molecule Sources view to open the Log view.
The Log view shows errors for all instances that could not be imported and warnings for any erronous but correctable entries in the original file. The most common errors are unsupported elements as the underlying NAOMI library does not support compounds with metals.
4. Select the created set and scroll through it in the Visual Selection view.
The Visual Selection view allows instant browsing even through large sets, as all depictions are calculated on the fly. New subsets can be created by selecting molecules and pressing the create subset button.
5. Click the ![]() icon and add the Duplicates property.
icon and add the Duplicates property.
Arbitrary properties can be added to molecules in MONA. If you add instance properties like instance name or external SD file properties, you have to choose if you want to use the first or all found values. In the case of numerical values you may alternatively select to use the minimum, average or maximum value of all instance values.
6. Sort the set by the Duplicates property.
Sets may be sorted by any registered property. Sorting by the number of duplicates allows to quickly find all molecules that appear multiple times in the original file.
7. Click ![]() and then on any molecule in the set to view its instances.
and then on any molecule in the set to view its instances.
The Instance View shows the location and details for all instances of the selected molecule. Additionally all properties currently registered for this molecule are shown.
8. Select the Histogram view and choose the MW (molecular weight) property in the combo box.
Histograms can be created on the fly for any numerical molecule property. Clicking the bins highlights the exact number of molecules for this bin.
Task 2: How can I convert a large table of SMILES into multiple SD files?
The second task shows how to split a set into multiple subsets and how to export these into SD files. We start with a set of molecules already loaded and selected.
1. Click on ![]() and select to split the set into 10 sets sorted by the property
and select to split the set into 10 sets sorted by the property ![]() MW.
MW.
After the splitting completed you can look at the first few sets sorted by the MW property to check the MW ranges: The first set contains all molecules with MW 1 to 155, the second set contains all with MW 155 to 203 and so on.
2. Select the original set again and split it into 10 shuffled subsets.
Using shuffled splits allows to easily create training and test sets to test your methods.
3. Select the splitted subsets and change their color by right clicking on the selection and choosing ![]() change color.
change color.
Color and type are used throughout MONA to identify sets. Colors are always shown and may be set freely. A set type describes how the set was initially created. This allows to see if a set was created from a file or the result of a set or filter operation. Type information for sets are implicitly created and are only shown if show simplified set symbols in the Preferences are disabled.
4. Select the splitted subsets and the original set. Change to the ![]() Histogram view, select the MW Property and enable normalize Y axis.
Histogram view, select the MW Property and enable normalize Y axis.
Selecting multiple sets while in the Histogram View allows to compare properties of these sets. In this case the subsets have approximately the same distribution as the original set, which is what you would expect from randomly choosen subsets.
5. Select the subsets splitted by MW and click ![]() to export them to SD files.
to export them to SD files.
Sets may be exported to either MOL, SD or SMILES files. Most often only the filename needs to be chosen. Filenames for multiple sets are automatically created by inserting _1, _2,... before the suffix. By default instances from all sources are selected. Choosing only some of the sources reduces the possible recreated instances to these sources. Either the first or all found instances for each molecule are written to the file.
Task 3: How to filter out compounds with undesired properties?
This task shows how to use the Filter view and set operations to create filtered subsets in MONA.
1. Create a set from the LigandExpo source by clicking ![]() in the Molecule Sources view and rename it by double clicking its name.
in the Molecule Sources view and rename it by double clicking its name.
Molecule Sources manages all file sources imported into MONA. Sets from file sources may be recreated any time.
2. Switch to the Filter view and create a new property filter ![]() that includes all molecules with MW between 200 and 400.
that includes all molecules with MW between 200 and 400.
All filters are expandable by clicking in the left corner. Expanded property filters show a histogram of the selected property for the currently selected set. Filter ranges may directly be specified by clicking and dragging in the histogram.
3. Click “Create Filtered Set” to apply the filter to the currently selected set.
If filters are sufficiently easy to evaluate (they contain no smarts filters and no tolerance) the number of molecules that fulfill the filter is interactively shown in the top right corner.
4. Create a functional group filter ![]() that includes all molecules containing a ketone group.
that includes all molecules containing a ketone group.
Functional group filters are much faster than SMARTS filters as the information that a molecule contains a group is precalculated upon import.
5. Create a chemical element filter ![]() excluding all molecules containing halogens and subtract
excluding all molecules containing halogens and subtract ![]() it from the LigandExpo set.
it from the LigandExpo set.
To create a set with molecules containing at least one halogen you first have to create a set with all molecules containing no halogens. By subtracting this set from the set with all molecules you get a set with molecules that contain at least one halogen. Alternatively you may apply a separate chemical element filter for each halogen and uniting the resulting sets.
6. Apply a property filter that includes all molecules containing at least 2 Donors.
7. Intersect ![]() the filtered sets to get a set with molecules fulfilling all filters.
the filtered sets to get a set with molecules fulfilling all filters.
In this case 217 molecules in the LigandExpo are in all filtered sets. Alternatively you may unite the sets to get all molecules fulfilling at least one filter. Or you may specify all filters as one chain and change the minimum number of filters that have to match in the top right corner.
Task 4: How do I cluster molecules by similarity?
Working with all molecules from the DrugBank 4.1 database we show how to cluster these molecules by tautomeric form and by similarity.
1. Add the Tautomer Identifier property to the set by clicking ![]() .
.
Tautomer and Protomer identifiers are binary properties that can only be used for sorting and clustering as their string representation has no meaning for humans.
2. Cluster ![]() the DrugBank set by the Tautomer Identifier property.
the DrugBank set by the Tautomer Identifier property.
Sets can have only one clustering attached at any time. Clusterings for properties either group by bin equal values for integer and string properties or by approximately equal values for real valued properties.
3. Switch the Visual Selection view to cluster ![]() mode and create a set with all molecules having more than one tautomer.
mode and create a set with all molecules having more than one tautomer.
Cluster mode shows the different clusters vertically starting with the largest cluster. The first 100 cluster members are shown by double clicking a cluster and scrolling horizontally. Sets from one or multiple clusters may be created by selecting the clusters and clicking create subset.
4. Select the set with all tautomers and use the anchor tool ![]() to align the depictions.
to align the depictions.
The alignment of 2D molecule depictions helps in spotting differences between similiar molecules. In this case it helps comparing which double bonds switch their positions from one tautomer to the other.
5. Cluster ![]() the DrugBank set by similarity with the parameters 100 clusters, 7 rounds, Euclidean similarity measure and Simple ECFP fingerprint.
the DrugBank set by similarity with the parameters 100 clusters, 7 rounds, Euclidean similarity measure and Simple ECFP fingerprint.
Similarity clustering employs a k-means algorithm which requires the number of expected clusters and the maximum number of rounds as parameters. If you want to cluster the molecules by similar depictions the Euclidean similarity together with the Simple ECFP fingerprint works best in our experience. Note that simple ECFP only considers the most basic graph topological information of the molecules and contains no chemical information (like the chemical elements or the bond order).
6. Create a new subset from a cluster and use the anchor ![]() tool to inspect the molecules in this set.
tool to inspect the molecules in this set.
As before the alignment of molecule depictions works best if the molecules in a set are similar to each other.
Task 5: How can I filter by a chemical pattern?
MONA supports filtering and searching for SMARTS patterns. In this task we start with the LigandExpo and search for all molecules containing a pteridine ring.
1. Search for a molecule containing a pteridine ring and use the pen tool ![]() to create a SMARTS pattern from this molecule.
to create a SMARTS pattern from this molecule.
Applying the SMARTS pattern should highlight the same molecule the pen was used on. You may freely edit the SMARTS expression in the text input box.
2. Click ![]() in the SMARTS input box to open the SMARTSEditor and delete all parts of the SMARTS except the pteridine ring.
in the SMARTS input box to open the SMARTSEditor and delete all parts of the SMARTS except the pteridine ring.
The SMARTSEditor can also be used to create arbitrarily complex SMARTS patterns from scratch.
3. Applying the SMARTS pattern highlights all matches in molecules in green and tries to align all other depictions to this pattern.
Note that only the first match is highlighted while multiple matches may be present. Another caveat are the hightlighted bonds: Internally the SMARTS match only consists of an atom match so the patterns CCCCCC and C1CCCCC1 lead to the same bond highlighting for a six membered ring.
4. Click “Create Set with Matches” to apply the current SMARTS pattern as filter that creates a new subset with all matches.
This leads to exactly the same result as creating a SMARTS filter in the Filter view which includes all molecules matching the pattern.
People and references
Mona has been developed by Matthias Hilbig and Sascha Urbaczek in the research group of Prof. Matthias Rarey at the Center for Bioinformatics of the University of Hamburg.
Please cite Mona with:
Hilbig, M.; Rarey, M. MONA 2: A Light Cheminformatics Platform for Interactive Compound Library Processing. J Chem Inf Model 2015, 55 (10), 2071-2078. DOI: https://doi.org/10.1021/acs.jcim.5b00292
Hilbig, M.; Urbaczek, S.; Groth, I.; Heuser, S.; Rarey, M. MONA - Interactive Manipulation of Molecule Collections. J Cheminform 2013, 5 (1), 38. DOI: https://doi.org/10.1186/1758-2946-5-38
