Our research group is dedicated to fundamental research in molecular biology, with the goal of understanding cellular and regulatory processes through data-driven approaches. We analyze large molecular datasets – often in national and international collaborations – such as those from RNA sequencing (bulk and single-cell), addressing fundamental questions on gene regulation, cell type identity, molecular networks, and disease mechanisms in health, aging, and disease.
Example: High-throughput single-cell and spatial transcriptomics
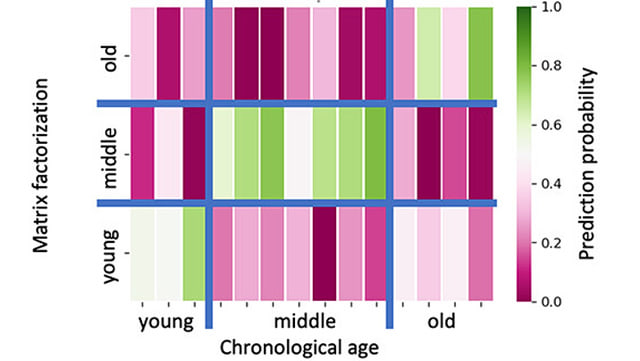
The group focuses on the analysis of high-dimensional molecular data generated by modern high-throughput technologies, in both bulk and single-cell experiments. In particular, we study gene and miRNA expression profiles to better understand biological processes at the transcriptional level. Bulk RNA sequencing provides robust averages across cell populations and is a well-established tool for analyzing differentially expressed genes and regulatory miRNAs under various biological conditions. Complementary to this, single-cell and single-nucleus RNA sequencing allow the investigation of individual cells, offering insights into cellular heterogeneity, rare cell types, and dynamic gene expression patterns at single-cell resolution. We have also gained experience in the emerging field of spatial omics technologies, which enable the integration of spatial and temporal coordinates with molecular measurements.
Example: Neurodegeneration and infectious disease research
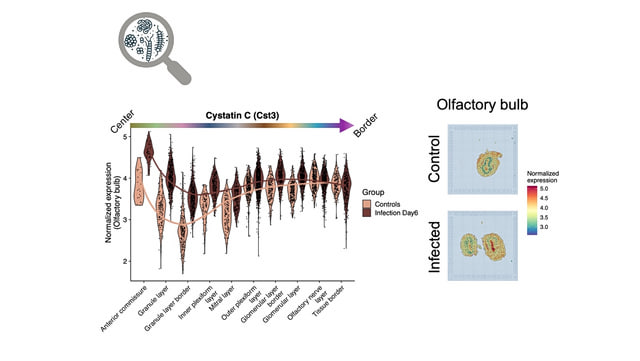
We are also engaged in the bioinformatic analysis of molecular biology data at the interface between neurodegenerative and infectious diseases. Our focus includes conditions such as Alzheimer’s disease, Parkinson’s disease, and viral infections, including those caused by SARS-CoV-2. The aim is to better understand the underlying molecular mechanisms and systematically capture disease-associated changes at the cellular and tissue levels.
For all these data types and modalities, we employ classical bioinformatics tools – such as quality control, normalization, and statistical analysis – together with modern machine learning and deep learning methods. These approaches enable us to detect complex patterns, identify cell types, reconstruct gene regulatory networks, and reveal disease-specific signatures. Such signatures allow us to describe disease processes at their root and systematically assess their translational potential for developing new biomarkers or targets for therapeutic intervention. By combining classical analytical methods with modern data-driven approaches, we contribute to advancing the understanding of age- and disease-related changes in the human brain and to elucidating immunological processes in infections.