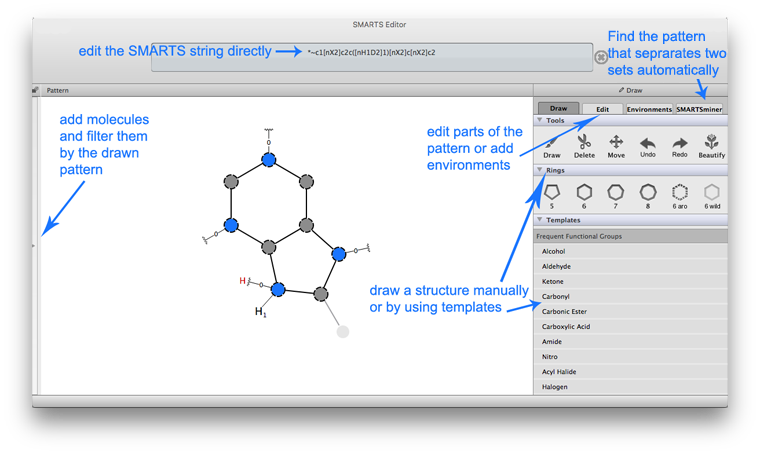
SMARTSeditor Documentation
SMARTSeditor - Common Tasks
The SMARTSeditor is a graphic editing tool for generic chemical patterns. Based on the SMARTS language, chemical patterns can be created and edited interactively, similar to molecule editing in a chemical structure editor. The following tasks show solutions for typical cheminformatics problems step by step.
Task 1:How do I generate a simple pattern representation based on a molecule structure?
The first task shows how to use the SMARTSeditor to generate a SMARTS by drawing a structure. In this case the example of acetic acid.
1. Click in the “Pattern“ area to create the first atom which is a carbon per default.
There are multiple ways to create an atom (all lead to identical results): Click in the SMARTS pattern text frame → type in the required element symbol (C in this example).
2. Add the three required atoms to the existing carbon by drag and drop out of the carbon atom.
In this case the drawn atom is carbon per default. It is necessary to edit the atom afterwards.
3. Edit the atoms by right clicking on the atom to change the element of the atoms.
It is also possible to switch to the “Edit” menu and select the atom you want to change. In this menu the element of an atom can be changed.
4. To edit the bonds quick edit them by right clicking and select the required bond type.
By using the “Edit” menu the bond type can also be changed.
5. Click on  to optimize the structure.
to optimize the structure.
The SMARTSeditor aligns the optimized structure to the drawn structure.
6. Afterwards copy the SMARTS string from the upper text frame to use it in other contexts.
To delete a pattern click on  .
.
Task 2: How can I add specific descriptions to my existing pattern?
The second task shows the different possibilities to add more descriptions to an exsiting SMARTS.
1. Create a pattern for purine by select  combine it with a second
combine it with a second  click on
click on  and delete one carbon from one ring. Close the ring with a new bond by clicking on and drag and drop the bond. Quick edit the bond by right clicking and select “Aromatic Bond”. Change the right carbons to nitrogens.
and delete one carbon from one ring. Close the ring with a new bond by clicking on and drag and drop the bond. Quick edit the bond by right clicking and select “Aromatic Bond”. Change the right carbons to nitrogens.
The SMARTSeditor has a part in the “Draw” menu in which rings were given as templates.
2. To add more information to the nitrogens edit them by right clicking ans select “Edit Atom”.
To edit the elements of a pattern it is also possible to enter the “Edit” menu and select an element of the pattern by clicking on it.
3. Add a common property by clicking on  in the “Common Properties” label. In the popped up menu select “Atomic Property”.
in the “Common Properties” label. In the popped up menu select “Atomic Property”.
The common properties which can be added were next to “Atomic Property” to add a “Chemical Element” or “Atomic Environment” to the selected atom.
4. Select the wanted property and increase the number if possible. Repeat this step until every nitrogen has its specific properties. After finishing the further information click on  to optimize the structure.
to optimize the structure.
The SMARTSeditor recognized if a selected property doesn't match to the valence and already selected properties and marks the SMARTS string if an error occurs.
5. Delete the pattern by clicking  . Afterwards go to the “Draw” menu and scroll through the templates until you find purine. Draw it by selecting it and click in the “Pattern” area.
. Afterwards go to the “Draw” menu and scroll through the templates until you find purine. Draw it by selecting it and click in the “Pattern” area.
The SMARTSeditor has many templates already given. Frequent used side chains, functional groups and ringsystems can be drawn by using the templates. It is afterwards possible to edit those templates until the description corresponds to the wanted pattern.
Task 3: How can I describe in which environment an atom or a bond is given in a pattern?
The third task shows how to add and adjust an environment of a pattern.
1. We want to create a SMARTS which matches each phenyl ring which belongs to a phylalanine side chain. For this purposes select the phenylalanaine side chain from the templates and draw it in the “Pattern” area.
2. Select the carbon atom from the ring which is connected to carbon of the alanine and switch to the “Edit” menu. Add a common property by clicking on  in the “Common Properties” label. In the popped up menu select “Atomic Environment”. Click on to edit the environment.
in the “Common Properties” label. In the popped up menu select “Atomic Environment”. Click on to edit the environment.
By clicking on  the SMATRTSeditor displays the environment as a thumbnail in the “Edit” menu.
the SMATRTSeditor displays the environment as a thumbnail in the “Edit” menu.
3. Draw the carbon of the alanine and the wild card atom with all the properties to the environment like they were displayed in the pattern. Close the environmental view.
After switching to the environmental pattern the handling of drawing elements in the SMARTSeditor is working like before. After accomplishe the work on an environmental pattern you can go back to the main pattern just by clicking on the “Pattern” lable of the main frame.
4. The last step is to delete the carbon and the wild card atom in the main pattern by clicking on  and selecting both elements by mouse over and click on them.
and selecting both elements by mouse over and click on them.
Task 4: How can I check for a pattern which appears in one set of molecules but does not appear in a second set of molecules?
The fourth task shows how to use the SMARTSminer to find discriminative SMARTS patterns of two given sets of molecules generated by a SMARTS.
1. Open the “Molecule” window and click on  . Select the first set of molecules. Repeat this step with the right column and the second set of molecules.
. Select the first set of molecules. Repeat this step with the right column and the second set of molecules.
It is also possible to use the “Molecule” window to apply a SMARTS pattern to a molecule set. In this case you create a SMARTS you would like to check and afterwards click on  to filter the molecule set with respect to this SMARTS pattern.
to filter the molecule set with respect to this SMARTS pattern.
2. Select SMARTSminer menu. In this menu the positive and negative support is set. Set the positive support to 70 percent and the negative support to 30 percent. Afterwards click on “Start”.
In this menu the positive side can be selected and the generalization can be set.
3. The discriminative patterns the SMARTSminer was found were displayed in the SMARTSminer menu. The SMARTS is displayed and some important information were included. By double click on one pattern it appears in the “Pattern” menu and can be used for further applications.
By clicking on  it is possible to delete molecules from a set. For this purpose select the molecules which should be deleted.
it is possible to delete molecules from a set. For this purpose select the molecules which should be deleted.
People and references
SMARTSeditor has been developed by Lars Wetzer, David Seier, Stefan Bietz and Karen Schomburg in the research group of Prof. Matthias Rarey at the Center for Bioinformatics of the University of Hamburg.
Please cite the SMARTSeditor with:
Schomburg, K. T., Wetzer, L., Rarey, M. Interactive Design of generic chemical patterns, Drug Discov Today (2013)
http://dx.doi.org/10.1016/j.drudis.2013.02.001
For further reading on the new SMARTSminer feature please see:
Bietz, S. Schomburg, K. T. Hilbig, M. Rarey, M. (2015). Discriminative Chemical Patterns: Automatic and Interactive Design. Journal of Chemical Information and Modeling, 55 (8), 1535ff.
http://pubs.acs.org/doi/abs/10.1021/acs.jcim.5b00323