New Paper: Galileo: Three-dimensional searching in large combinatorial fragment spaces on the example of pharmacophores
22. Dezember 2022
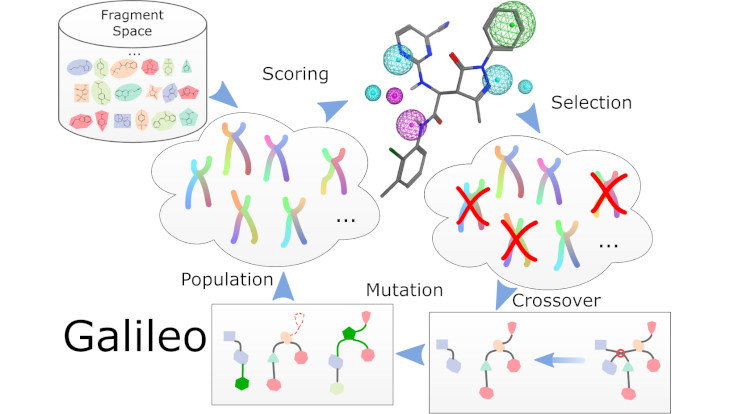
Foto: Christian Meyenburg
Chemical fragment spaces contain billions of molecules. This makes the process of searching them for bioactive compounds computationally challenging. However, searching is not necessarily equal to screening. While search algorithms on the level of molecular topology have been presented already, searching with 3D queries like molecular superposition, docking or pharmacophore mapping remains challenging. In the paper, we presented a genetic algorithm called Galileo, that operates exactly on a given fragment space (e.g. Enamine's REAL Space). It is able to handle arbitrary scoring functions that can be attached as external software modules. Additionally, Galileo contains a pharmacophore mapping tool called Phariety which enables pharmacophore searches in fragment spaces. For all details, have a look at https://doi.org/10.1007/s10822-022-00485-y. The new software tools Galileo and Phariety can be found in our NAOMI ChemBio software collection at https://uhh.de/naomi

