New Paper: Searching Geometric Patterns in Protein Binding Sites and Their Application to Data Mining in Protein Kinase Structures
15. September 2021
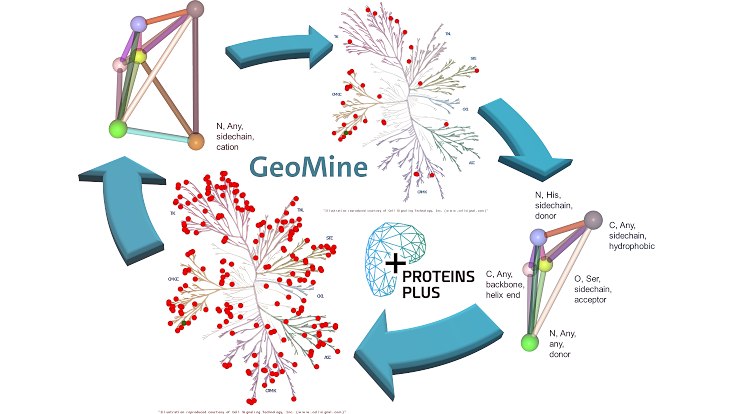
Foto: Figure: The human kinome trees in this figure were created with KinMap (http://www.kinhub.org/kinmap/)
Protein kinase research encounters several challenges, the most serious one being the improvement of inhibitor selectivity through compound optimization and covalent inhibitor design. In our recent publication, we face these challenges by applying data mining in the Protein Data Bank through the tool GeoMine. It is an easily accessible tool on the ProteinsPlus web server (https://proteins.plus) which enables user-defined similarity searches in binding sites within minutes. The automated screening for interaction- and property-based similarities offers a powerful tool to medicinal chemists. Especially in the field of kinase inhibitor design, this approach comes in handy as idea generator for advancing known kinase inhibitors and learning from the already available structural knowledge of protein kinase-ligand complexes. The summary of the tool's capabilities and three use cases in the full paper (https://pubs.acs.org/doi/10.1021/acs.jmedchem.1c01046) provide a comprehensive introduction to using GeoMine for kinase inhibitor design.