LTRdigest
is a software tool for automated annotation of internal features of LTR retrotransposons.
Background
LTR retrotransposons and endogenous retroviruses (ERVs) are transposable elements in eukaryotic genomes well suited for computational identification. De novo identification tools such as LTRharvest determine the position of potential LTR retrotransposon or ERV insertions in genomic sequences. For further analysis, it is desirable to obtain an annotation of the internal structure of such candidates. LTRdigest extends the results of current de novo LTR retrotransposon prediction tools. It is able to identify protein coding regions associated with the retrotransposition process as well as internal regulatory features using local alignment and hidden Markov model-based algorithms.
It accepts sequence data in a variety of formats and a basic LTR annotation in GFF3format as input. Protein domains are specified as profile HMMs in HMMER3 format. As output, LTRdigest creates GFF3 and CSV annotations as well as FASTA sequence files for all detected features.
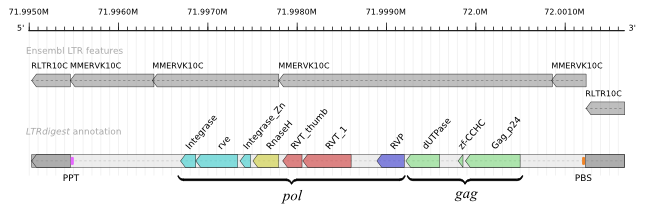
Figure: Example annotation of an LTR retrotransposon candidate from Mus musculus chromosome 4, positions 71,995,000-72,001,670. The top track shows sequence-based LTR retrotransposon matches from Ensembl release 54 (May 2009). LTR matches and internal region matches can only be linked by their Repbase feature identifier ("RLTR10C" for the LTR matches and "MMERVK10C" for internal region matches). The bottom track contains a representation of a hierarchical LTR retrotransposon annotation graph, as reported by LTRdigest, collapsed into a single track. The grey blocks at the end of the element represent the LTRs, while the coloured elements in the middle represent protein domain hits. The orange and purple blocks at the inner LTR boundaries represent PBS and PPT features, respectively.
For a detailed description of the software see the manual. An independent source of information can be found in the blog of Avril Coghlan.
Applications
We used LTRdigest to annotate the chromosome 4 sequence of Mus musculus (Results). These results were used to identify 88 (near) full-length ERVs (Results), separating them from truncated insertions and other repeats.
We also performed an example annotation and classification of LTR retrotransposon insertions in the Drosophila melanogaster 5.8 genome as a proof of concept. Complete annotations as well as the members and sequences of the 62 groups reported are available for download.
Availability
The source code is freely available as part of GenomeTools which is released under a BSD-like open source license. Please get the latest stable source or binary distribution from the GenomeTools web site.
LTRdigest is also currently available in the Debian unstable and Ubuntu raring and saucy package repositories (genometools package) as part of Debian Med.
Developers
Sascha Steinbiss
Ute Willhoeft
Gordon Gremme
Stefan Kurtz
Publication
S. Steinbiss, U. Willhoeft, G. Gremme and S. Kurtz:
Fine-grained annotation and classification of de novo predicted LTR retrotransposons.
Nucleic Acids Research, 37(21):7002-7013 (2009)
Contact
Sascha Steinbiss
Center for Bioinformatics, University of Hamburg
Bundesstr. 43, 20146 Hamburg, Germany
Phone +49 40 42838 7322, Fax. +49 40 42838 7312
