LTRsift
LTRsift is a graphical desktop tool for semi-automatic postprocessing of de novopredicted LTR retrotransposon annotations, such as the ones generated by LTRharvestand LTRdigest.
Its user-friendly interface displays LTR retrotransposon candidates, their putative families and their internal structure in a hierarchical fashion (see screenshot below), allowing the user to "sift" through the sometimes large results of de novo prediction software. It also offers customizable filtering and classification functionality.
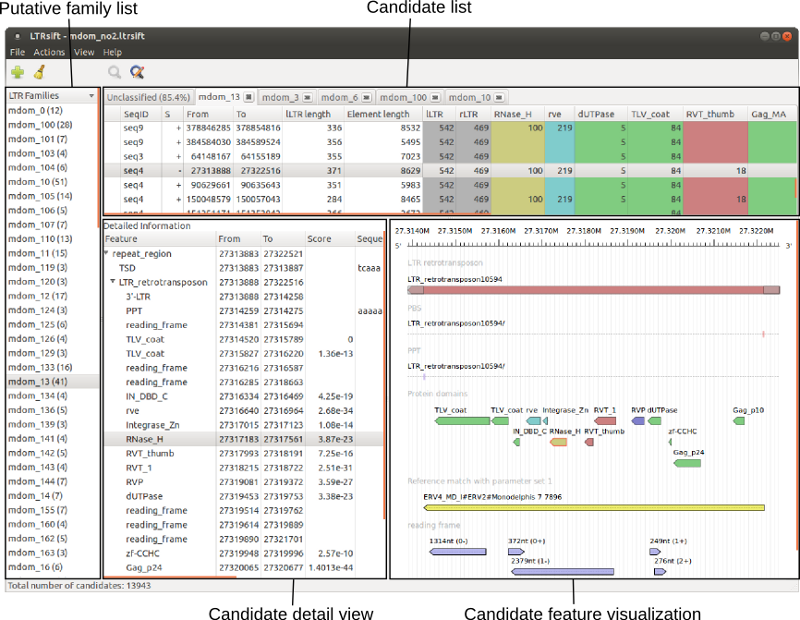
To ease manual curation it also supports GUI-driven reassignment, splitting and further annotation of candidates. Export of grouped candidate sets is possible in standard formats (GFF3 for annotations, FASTA for sequences).
Features
implements the classification scheme proposed in:
S. Steinbiss et al.
Fine-grained annotation and classification of de novo predicted LTR retrotransposons.
Nucleic Acids Research, 37(21):7002–7013 (2009).
wizard-guided project creation and automated pre-classification (LASTrequired)
"drag-and-drop'' manual family assignment
candidate filtering based on dynamic, customizable rule sets, defined in a simple programming language
reference matching against custom FASTA files (BLAST required)
ORF detection
concise, customizable visualization of candidates using AnnotationSketch
candidate lists can be sorted by column values
all project data stored in one persistent database
Availability
LTRsift can be downloaded here for 64-bit Intel Linux and Mac OS X platforms.
LTRsift v1.0.2 (Linux 64-bit)
LTRsift v1.0.2 (Mac OS X 64-bit)
The source code (GPL) is available from a Git repository at http://github.com/satta/ltrsift.
Note that some external libraries, e.g. Cairo and GTK+, are required. Read more about these dependencies.
LTRsift is also currently available in the Debian unstable and Ubuntu package repositories (packages ltrsift and ltrsift-examples) as part of Debian Med.
Bug reports and wishlist
Have you encountered something that you feel may be a bug in LTRsift or do you have a suggestion for improvement? Just file a ticket in our on-line issue tracker!
Example Data
We offer input GFF3 candidate annotations from the paper (from LTRharvest/LTRdigest) and the corresponding sequences in FASTA format for download as gzipped tar archives:
Drosophila melanogaster use case (39 MB)
Monodelphis domestica use case (947 MB)
Developers
LTRsift has been developed by Sascha Kastens and Sascha Steinbiss, with parts of the software written by Joachim Bonnet.
Publication
S. Steinbiss, S. Kastens and S. Kurtz:
LTRsift: a graphical user interface for semi-automatic classification and postprocessing of de novo detected LTR retrotransposons.
Mobile DNA, 3:18 (2012)
Contact
Sascha Steinbiss
Center for Bioinformatics, University of Hamburg
Bundesstr. 43, 20146 Hamburg, Germany
Phone +49 40 42838 7322, Fax. +49 40 42838 7312
